BlobTools Workflows
Summary
BlobTools offers two main workflows for taxonomic interrogation of paired-end (PE) read datasets depicted in the figure below.
- Workflow A: targeted at de novo genome assembly projects without a reference genome
- Workflow B: targeted at re-sequencing projects where a reference genome is available
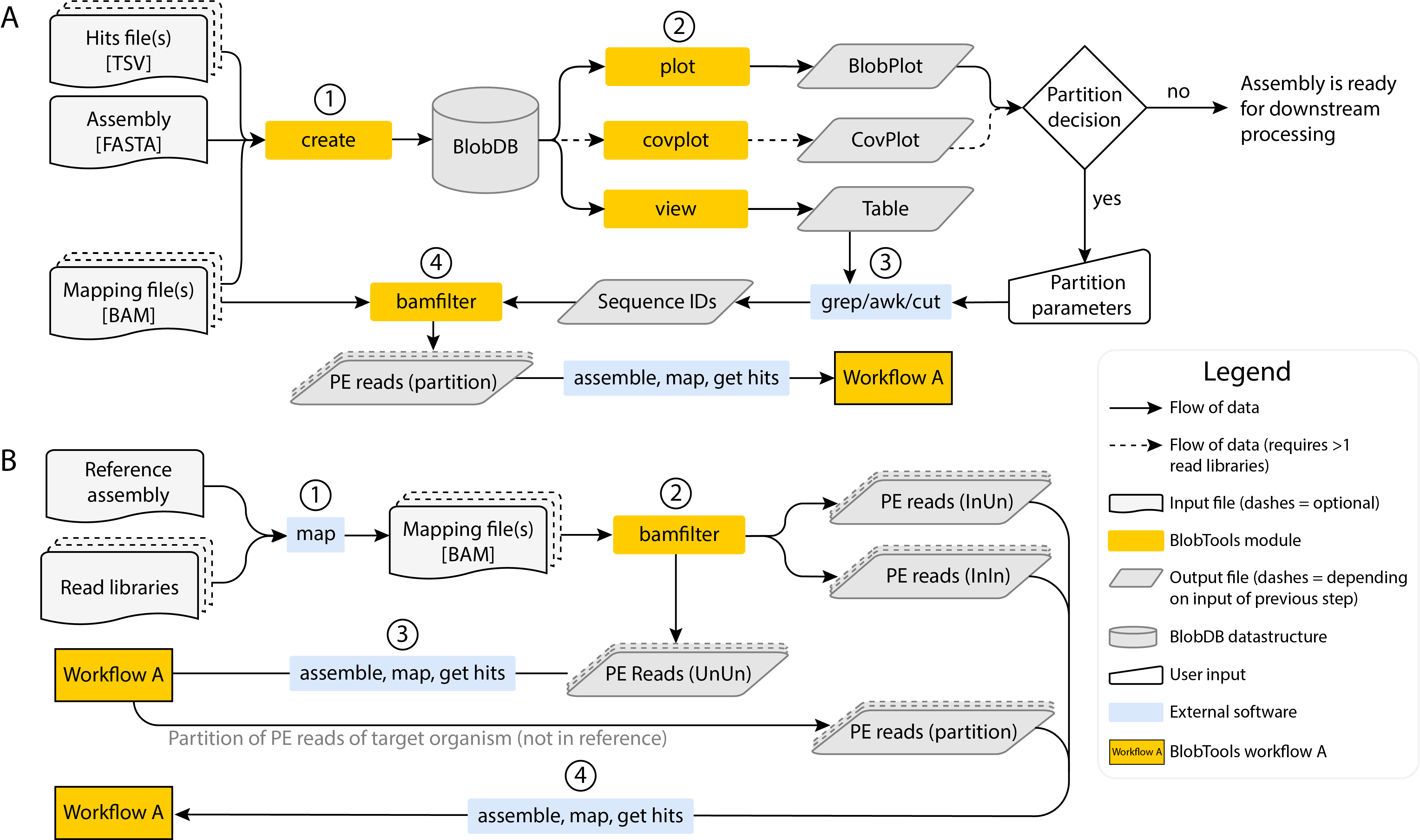
Main BlobTools workflows for taxonomic interrogation of paired-end (PE) read datasets. A: Workflow A. B: Workflow B
Workflow A
Steps
- Construction a BlobDB data structure based on input files
- Visualisation of assembly and generation of tabular output
- Partitioning of sequence IDs based on user-defined parameters informed by the visualisations
- Partitioning of paired-end reads based on their mapping behaviour to sequence partitions
Resulting reads are then assembled by partition and the assemblies can be screened again using the workflow.
Comments
- Although the BlobTools module
create(step A1) supports multiple mapping formats, it is recommended that these are processed in advance usingmap2covto reduce file size. - Module
bamfilter(step A4) is only of relevance if paired-end read data is used, since single-end read data can easily be partitioned usingGNU grepor other tools.
Workflow B
Steps
- Reads are mapped against the reference genome
- Resulting BAM file is processed with the module
bamfilterusing the parameter--include_unmappedand without providing a lists of sequences. This will result in three FASTQ files: InIn, InUn and UnUn (see mapping behaviour) - Since taxonomic origin of the InIn and InUn reads has been established through the mapping step, only the UnUn reads are assembled de novo and processed via workflow A. This decreases computational requirements substantially.
- If workflow A yields a paired-end read partition of the target organism, which will consist of parts of the organism’s genome not present in the reference, these reads are can be used together with the InIn and InUn reads (step B2) to generate a new assembly, which should be screened again via workflow A
Comments
- This iterative procedure can easily be applied to projects studying highly variable species where segmental presence-absence is common and a reference genome is expanded (to form a pangenome) as new samples are sequenced, or holobiomes, where reference genomes of multiple taxa are expanded as new samples are added
Updated over 8 years ago
