My first blobplot
This section describes the how to generate a BlobPlot based on the files provided in the example/ directory.
1. Create a blobDB file from input files
1.1 Input file requirements
- one assembly file, e.g.
example/assembly.fna - one (or more) coverage file(s) e.g.
example/mapping_1.bam - one (or more) hits file(s), e.g.
example/blast.out
1.2 Creating a blobDB
# using the files provided in the test_files/ folder
./blobtools create \
-i example/assembly.fna \
-b example/mapping_1.bam \
-t example/blast.out \
-o example/my_first_blobplot
2. Create a view of a blobDB file
2.1. Creating a view
- Using blobtools
view, information stored in a blobDB can be extracted. - Using the default settings, blobtools
viewwill generate a tabular output for the taxonomic rank of "phylum".
./blobtools view \
-i example/my_first_blobplot.blobDB.json \
-o example/
2.2. Inspecting the output
- The resulting file can be inspected using a one-liner (to indent the columns properly), e.g.:
grep '^##' example/my_first_blobplot.blobDB.table.txt ; \
grep -v '^##' example/my_first_blobplot.blobDB.table.txt | \
column -t -s $'\t'
## blobtools v1.0
## assembly : example/assembly.fna
## coverage : bam0 - example/mapping_1.bam
## taxonomy : tax0 - example/blast.out
## nodesDB : data/nodesDB.txt
## taxrule : bestsum
##
# name length GC N bam0 phylum.t.6 phylum.s.7 phylum.c.8
contig_1 756 0.2606 0 90.406 Actinobacteria 200.0 0
contig_2 1060 0.2623 0 168.409 Actinobacteria 2300.0 0
contig_3 602 0.2342 0 43.761 Actinobacteria 10000.0 0
contig_4 951 0.3155 0 456.313 Actinobacteria 1000.0 0
contig_5 614 0.329 0 163.557 Nematoda 2000.0 0
contig_6 216 0.1944 0 25.88 Tardigrada 4000.0 2
contig_7 4060 0.2584 0 52.312 Nematoda 2000.0 0
contig_8 2346 0.2801 0 91.742 unresolved 2000.0 1
contig_9 1599 0.2439 0 74.757 Nematoda 200.0 0
contig_10 6273 0.3067 0 310.634 no-hit 0.0 None
- The main header (##) contains version number, input files and parameters used while creating the blobDB file.
- The table header (#) indicates the names of the columns (explained in the table below).
- All following rows contain information about the sequences in the order they appear in the assembly file.
| column header | description |
|---|---|
| name | name of the sequence |
| length | total length of the sequence, i.e. count(A, G, C, T, N) |
| GC | GC content percentage of the sequence, i.e. count(G, C)/count(A, G, C, T) |
| N | Number of N's in the sequence, i.e. count(N) |
| bam0 | Coverage from bam0 (see main header for filename) |
| phylum.t.6 | The assigned taxonomy of the sequence at the taxonomic rank of "phylum" under the tax-rule "best-sum" |
| phylum.s.7 | The sum of scores for the taxonomy of the sequence at the taxonomic rank of "phylum" under the tax-rule "best-sum" |
| phylum.c.8 | The c-index for the taxonomy of the sequence at the taxonomic rank of "phylum" under the tax-rule "best-sum" |
- If N > 1 coverage files are provided, N+1 coverage columns would show up in the table. One column per coverage files, and an additional column listing the sum of coverages across all files.
- The last three columns list information about the taxonomy at the taxonomic rank of "phylum" under the tax-rule "best-sum".
- If N > 1 taxonomic ranks are specified using the
-rargument, additional columns will appear in the table. - If the
--hitsflag is used, the table will include the sum of scores by hits file for each taxID - The headers of the "taxonomy" columns are numbered (1-based) to ease filtering using command line applications such as
GNU awkorUNIX/LINUX cut.
3. Create a blobplot
./blobtools plot \
-i example/blobDB.json \
-o example/
- This generates three files:
- the blobplot
blobDB.json.bestsum.phylum.p7.span.100.blobplot.bam0.png - the readcovplot
blobDB.json.bestsum.phylum.p7.span.100.blobplot.read_cov.bam0.png - the stats file
blobDB.json.bestsum.phylum.p7.span.100.blobplot.stats.txt
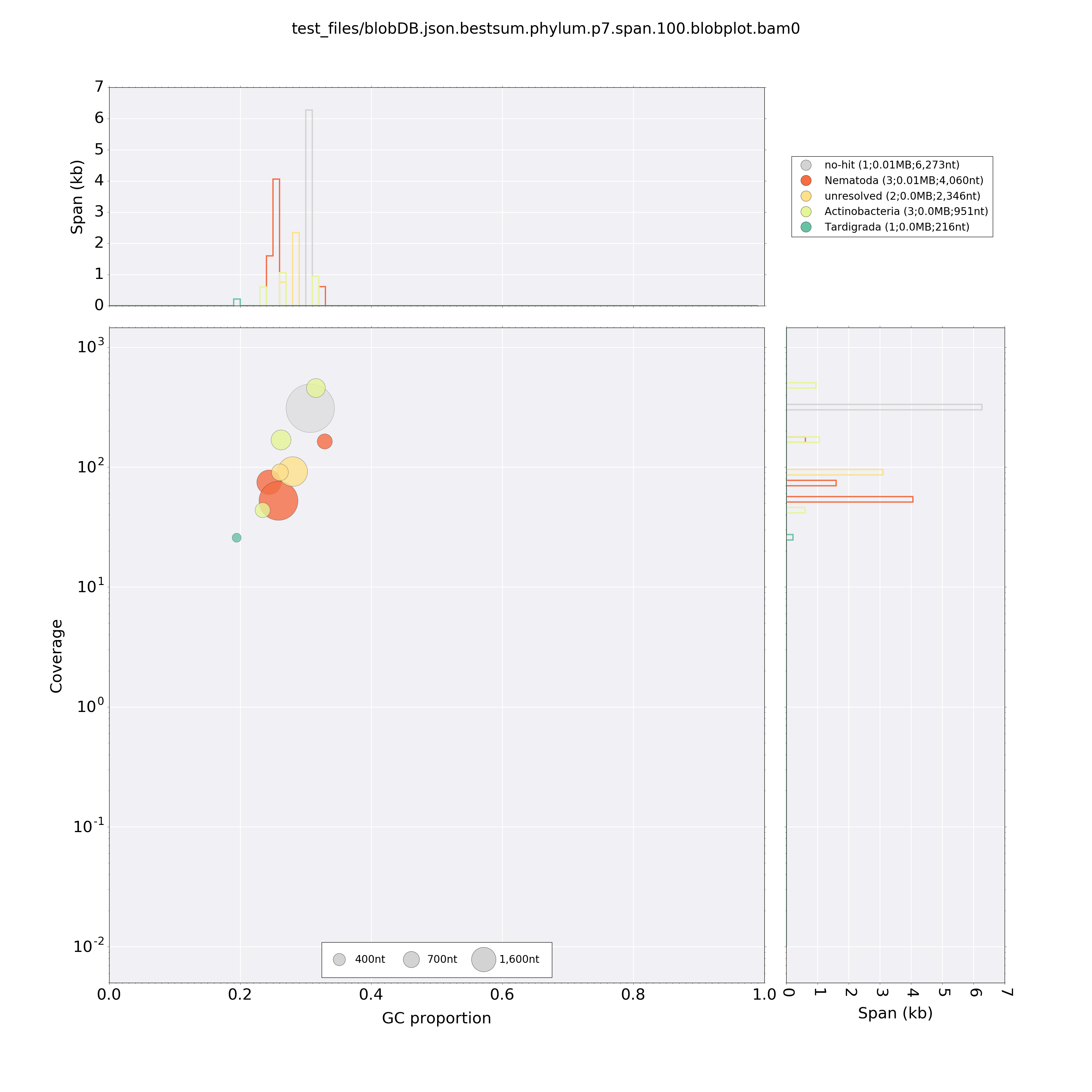
BlobPlot of example/blobDB.json
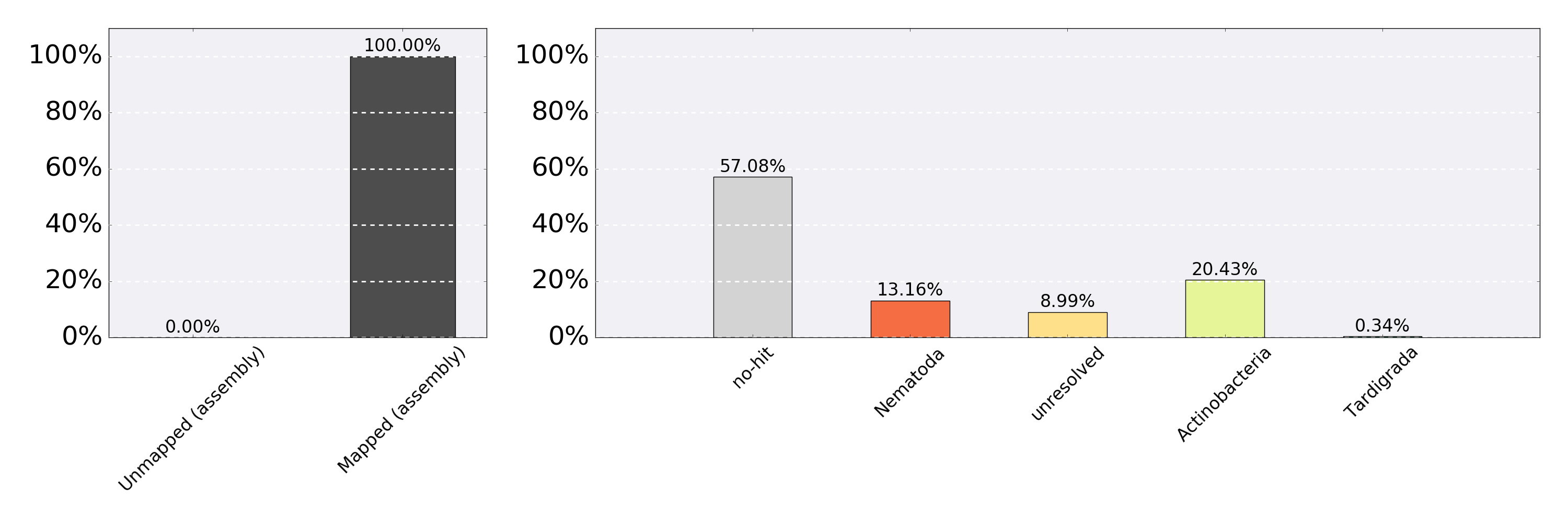
ReadCovPlot of example/blobDB.json
Updated over 8 years ago
