BlobPlot
Description
- BlobPlots are two-dimensional scatter plots, decorated with coverage and GC histograms
- Sequences are represented by circles (with diameters proportional to sequence length) in the scatter plot and coloured by taxonomic affiliation
- Circles are positioned on the Y-axis in the scatter plot based on the base coverage of the sequence in the coverage library, a proxy for molarity of input DNA in the sequencing reaction
- Circles are position on the X-axis based on their GC content, the proportion of G and C bases in the sequence, which can differ substantially between genomes
- Coverage and GC histograms are drawn for each taxonomic group, which are weighted by the total span (cumulative length) of sequences occupying each bin
- A legend reflects the taxonomic affiliation of sequences and lists count, total span and N50 by taxonomic group
- Taxonomic groups can be plotted at any taxonomic rank and colours are selected dynamically from a colour map
- The number of taxonomic groups to be plotted can be controlled (
--plotgroups, default is ‘7’) and remaining groups are binned into the category ‘others’
Example
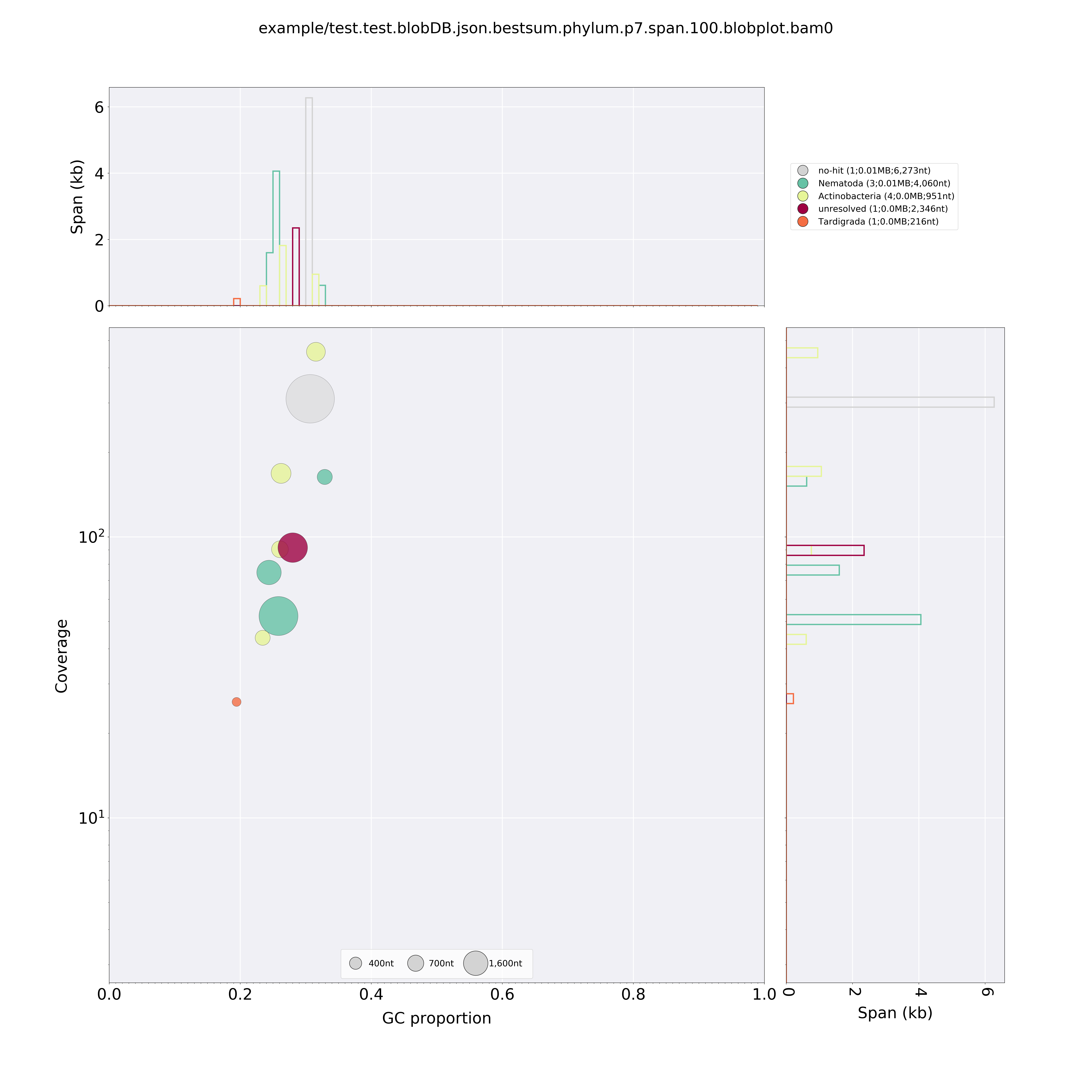
BlobPlot of example/blobDB.json
Updated over 8 years ago
